| dyads_test_vs_ctrl_m2_shift8 (dyads_test_vs_ctrl_m2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m2; m=0 (reference); ncol1=16; shift=8; ncol=24; --------dhtGmCACGTGkCarh
; Alignment reference
a 0 0 0 0 0 0 0 0 78 65 44 28 167 9 244 6 11 8 8 14 27 107 74 68
c 0 0 0 0 0 0 0 0 44 67 56 16 67 219 1 229 4 9 5 11 186 52 53 65
g 0 0 0 0 0 0 0 0 68 50 48 183 8 10 8 4 234 0 237 156 14 54 69 53
t 0 0 0 0 0 0 0 0 64 72 106 27 12 16 1 15 5 237 4 73 27 41 58 68
|
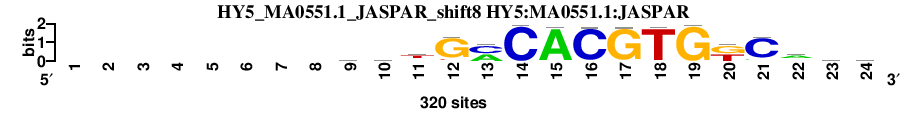
| HY5_MA0551.1_JASPAR_shift8 (HY5:MA0551.1:JASPAR) |
 |
0.973 |
0.973 |
7.904 |
0.973 |
0.989 |
6 |
1 |
15 |
1 |
4 |
5.400 |
1 |
; dyads_test_vs_ctrl_m2 versus HY5_MA0551.1_JASPAR (HY5:MA0551.1:JASPAR); m=1/115; ncol2=16; w=16; offset=0; strand=D; shift=8; score= 5.4; --------wwtGmCACGTGkCaww
; cor=0.973; Ncor=0.973; logoDP=7.904; NsEucl=0.973; NSW=0.989; rcor=6; rNcor=1; rlogoDP=15; rNsEucl=1; rNSW=4; rank_mean=5.400; match_rank=1
a 0 0 0 0 0 0 0 0 103 108 52 9 150 0 310 3 1 0 2 4 24 183 94 89
c 0 0 0 0 0 0 0 0 60 56 55 8 165 317 1 311 5 9 1 1 279 30 62 68
g 0 0 0 0 0 0 0 0 68 62 30 279 1 1 9 5 311 1 317 165 8 55 56 60
t 0 0 0 0 0 0 0 0 89 94 183 24 4 2 0 1 3 310 0 150 9 52 108 103
|
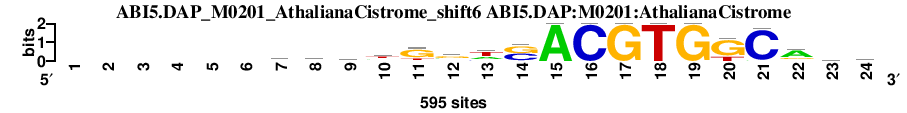
| ABI5.DAP_M0201_AthalianaCistrome_shift6 (ABI5.DAP:M0201:AthalianaCistrome) |
 |
0.872 |
0.775 |
7.904 |
0.947 |
0.955 |
54 |
4 |
16 |
25 |
34 |
26.600 |
16 |
; dyads_test_vs_ctrl_m2 versus ABI5.DAP_M0201_AthalianaCistrome (ABI5.DAP:M0201:AthalianaCistrome); m=16/115; ncol2=18; w=16; offset=-2; strand=D; shift=6; score= 26.6; ------wdrwgrwsACGTGKCarw
; cor=0.872; Ncor=0.775; logoDP=7.904; NsEucl=0.947; NSW=0.955; rcor=54; rNcor=4; rlogoDP=16; rNsEucl=25; rNSW=34; rank_mean=26.600; match_rank=16
a 0 0 0 0 0 0 241 217 224 161 44 157 198 0 595 0 0 0 0 0 27 394 151 190
c 0 0 0 0 0 0 71 59 80 39 19 119 92 289 0 595 0 0 0 0 568 50 124 117
g 0 0 0 0 0 0 131 167 158 98 400 296 0 294 0 0 595 0 595 446 0 123 174 87
t 0 0 0 0 0 0 152 152 133 297 132 23 305 12 0 0 0 595 0 149 0 28 146 201
|
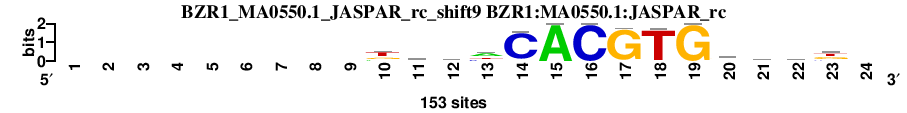
| BZR1_MA0550.1_JASPAR_rc_shift9 (BZR1:MA0550.1:JASPAR_rc) |
 |
0.888 |
0.777 |
7.182 |
0.948 |
0.962 |
46 |
3 |
40 |
24 |
28 |
28.200 |
19 |
; dyads_test_vs_ctrl_m2 versus BZR1_MA0550.1_JASPAR_rc (BZR1:MA0550.1:JASPAR_rc); m=19/115; ncol2=14; w=14; offset=1; strand=R; shift=9; score= 28.2; ---------kkbhCACGTGkmkk-
; cor=0.888; Ncor=0.777; logoDP=7.182; NsEucl=0.948; NSW=0.962; rcor=46; rNcor=3; rlogoDP=40; rNsEucl=24; rNSW=28; rank_mean=28.200; match_rank=19
a 0 0 0 0 0 0 0 0 0 33 28 16 72 0 153 0 0 4 0 12 51 24 0 0
c 0 0 0 0 0 0 0 0 0 0 22 43 39 142 0 153 0 0 0 26 52 27 30 0
g 0 0 0 0 0 0 0 0 0 39 61 53 0 5 0 0 147 2 153 66 20 47 60 0
t 0 0 0 0 0 0 0 0 0 81 42 41 42 6 0 0 6 147 0 49 30 55 63 0
|
| PIF3_PIF3_2_Athamap_shift8 (PIF3:PIF3_2:Athamap) |
 |
0.830 |
0.782 |
8.481 |
0.938 |
0.939 |
88 |
2 |
2 |
34 |
59 |
37.000 |
28 |
; dyads_test_vs_ctrl_m2 versus PIF3_PIF3_2_Athamap (PIF3:PIF3_2:Athamap); m=28/115; ncol2=17; w=16; offset=0; strand=D; shift=8; score= 37; --------rdrvCCACGTGGvmvs
; cor=0.830; Ncor=0.782; logoDP=8.481; NsEucl=0.938; NSW=0.939; rcor=88; rNcor=2; rlogoDP=2; rNsEucl=34; rNSW=59; rank_mean=37.000; match_rank=28
a 0 0 0 0 0 0 0 0 15 9 10 8 1 0 32 0 0 0 0 0 9 15 11 7
c 0 0 0 0 0 0 0 0 2 4 2 11 26 32 0 32 0 0 0 0 10 15 13 8
g 0 0 0 0 0 0 0 0 9 10 13 13 5 0 0 0 32 0 32 32 11 0 8 11
t 0 0 0 0 0 0 0 0 6 9 7 0 0 0 0 0 0 32 0 0 2 2 0 6
|




